Search help
Table of Contents
Our search allows users to search all of our active gene symbol reports with ease. The search server utilises Apache Solr which offers a powerful full-text search and hit highlighting.
The search form can be found within the mast head of each page. Enter a search term within the search input box and click on the spy glass icon or press enter when the focus is on the input box.
Basic search
The simplest way to search is to type a query word/ID into the input box within the mast head and click on the spy glass. The default option is a full-text search over all the indexes and fields. If reports are found containing the keyword/ID they are displayed in order of relevance (for more information see indexed fields for each search type). Users can change the default number of results per page from 10 up to 50.
The results display specific fields from within the search index. The first line of each result contains the gene symbol and the gene name. The second row will show some of the important fields to help identify the hit. The third row reports the field the keyword/ID matches, so if the keyword matched an approved symbol within a gene symbol report the third row would say "Matches: Gene symbol etc" as seen below in figure 1.
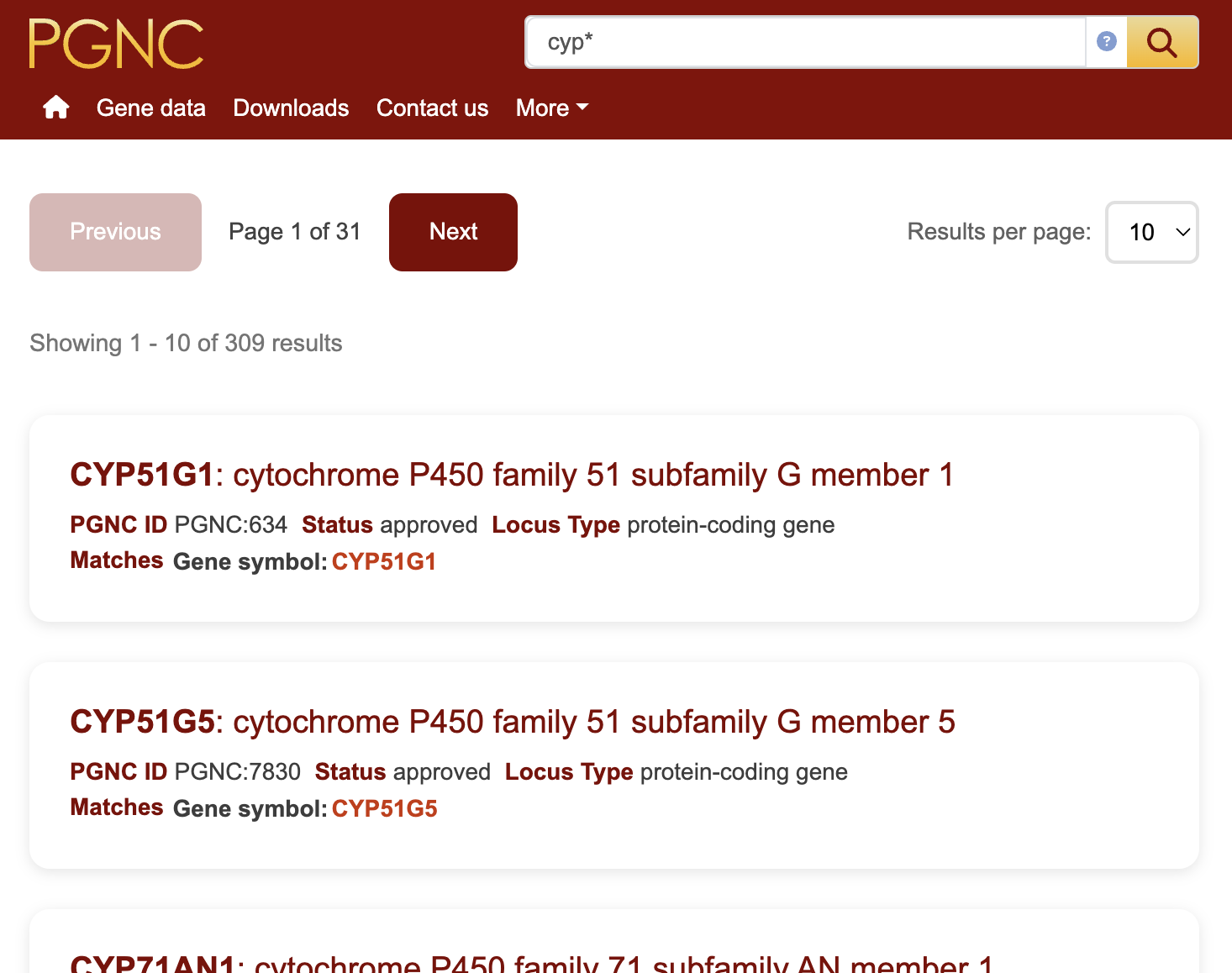
Advanced search
The search application allows users to make advanced queries using the search box. In this section we describe how to use wildcards, logic operators and specify indexed fields.
Wildcard search
Sometimes it may be useful to match records based on a query pattern rather than a keyword or ID. Our search allows users to use wildcard operators with an asterisk (*) to stand in place for one or more characters and a question mark (?) to stand in place for a single character substitution. Multple wildcards can be used in the same query, for instance searching for CYP?1* will find the symbols CYP51G1, CYP51G5, CYP71AN1 etc.
Logic operators
By default the search application uses the logical operator OR, so inputing CDC4* LFY into the search box actually equates to CDC4* OR LFY so one result will contain CDC4* and one will contain LFY. Sometime however this isn't what a user wants to search. Lets say a user wants to find reports that contain both CYP* and PGNC:634 in the same report. By changing the search query to CYP* AND PGNC:634 the results returned are more pertinent. Alternatively asking for reports that do not contain a keyword/ID may be preferable therefore users may use NOT or - within term such as CYP* NOT PGNC:634 or CYP* -PGNC:634.
Phrases
As discussed above the default operator is OR, so if a search using the term cytochrome P450 family 51 subfamily G member 5 was used the actual search term will be cytochrome OR P450 OR family OR 51 OR subfamily OR G OR member OR 5. This can be addressed by simply quoting the query so that the search knows to treat the quoted block as one term like "cytochrome P450 family 51 subfamily G member 5".
Indexed fields
Users may search within a specific indexed field by using the information seen in the indexed fields section below using a very basic notation. To specify an index the user need only to type in to the search field the indexed field key followed immediately with a colon (:) and then the query eg ncbi_gene_id:7454686. If the query is not an ID or a keyword "phrases" can be used after the colon eg gene_name:"cell division cycle 45".
List of indexes
- alias_gene_name
- Other names used to refer to a gene as seen in the "Alias names" field in the gene symbol report
- alias_gene_symbol
- Other symbols used to refer to a gene as seen in the "Alias symbols" field in the gene symbol report
- chromosome
- Chromosome location of the gene
- ensembl_gene_id
- Ensembl gene ID.
- gene_name
- PGNC approved name for the gene. Equates to the "Approved name" field within the gene symbol report
- gene_symbol
- The PGNC approved gene symbol. Equates to the "Approved symbol" field within the gene symbol report
- locus_type
- The locus type (a.k.a biotype) as set by the PGNC
- primary_id
- The ID of the gene from the primary resource in which we used as a starting point for naming the gene
- ncbi_gene_id
- NCBI gene ID for the gene
- pgnc_id
- PGNC ID. A unique ID created by the PGNC for every approved symbol
- phytozome_id
- Phytozome is a plant comparative genomics portal, integrating plant genome sequences and annotation data. Currently in the database all of the Phytozome IDs begin with "Potri" as we currently only display Populus trichocarpa genes
- prev_gene_name
- Gene names previously approved by the HGNC for a gene. Equates to the "Previous names" field within the gene symbol report
- prev_gene_symbol
- Gene symbols previously approved by the HGNC for this gene. Equates to the "Previous symbols" field within the gene symbol report
- status
- Status of the symbol report, which can be either "Approved" or "Entry Withdrawn"
- taxon_id
- The NCBI taxonomy ID of the species of interest
- uniprot_id
- UniProt protein accessions for the protein products of the gene